Development of a set of 24 SNP (single nucleotide polymorphism) markers that allows 192 samples to be simultaneously analysed in a single day for 24 markers associated with agronomic traits of interest. SNP markers represent polymorphisms due to differences consisting in one nucleotide only, they are among the most useful and widely used for molecular characterisation. The EP1 Fluidigm instrument used for the analyses, thanks to its high throughput, allows it to be used for assisted breeding projects that entail the analysis of populations consisting of many individuals.
 Fluidigm EP1 Genotyping platform
Fluidigm EP1 Genotyping platform
SNP markers (single nucleotide polymorphism) are the most frequent type of genetic variation and evenly distributed across the whole genome. They feature advantages with respect to other molecular markers such as the presence of 2 alleles and transferability of protocols between laboratories, experiments, etc. The analysis platform may be applied to agrifood sector and offers to companies a genotype analysis and molecular selection service with rapid timing (required time from 2-3 weeks to one work day) and at low costs.
The chip features make it applicable to both horticultural and cereal crops. The laboratory is able to develop the sets of 24 SNP markers ( and multiples) associated to various characters of agronomic interest for any given plant species and provide genotyping and assisted selection services for 1-24 (and multiples of 24) markers. The service targets seed companies and breeders. A rapid and precise molecular characterisation is useful to speed up the genetic improvement process by companies.
 Chip used for analyses
Chip used for analyses
Set of 24 SNP markers associated with the most important agronomic traits of bread wheat
The set of 24 markers was developed upon demand by a seed sector company for the purpose of characterising a collection of common wheat of reference. To develop the chip, traits of agronomic interest were chosen on the company's request (resistance to disease, traits connected to quality etc.). Development unfolded in a first bibliographic research stage to assess genes associated to the traits of interest, which was followed by a bioinformatic analysis of sequences for identification of polymorphisms (SNP) responsible for allelic variants with improved features. After an optimisation stage that led to choosing the most suitable markers, a set of 24 locus specific and co-dominant markers was obtained. A collection of bread wheat of reference for the seed sector company was characterised with the 24 SNP markers. Thanks to the "chip" format used, the analysis was quick and reliable.
Company in the seed sector
The data of the SNP markers and more detailed phenotypic assessments, will allow the company to identify accessions with desired characteristics with regard to biotic and abiotic stress tolerance, wheat production and quality.
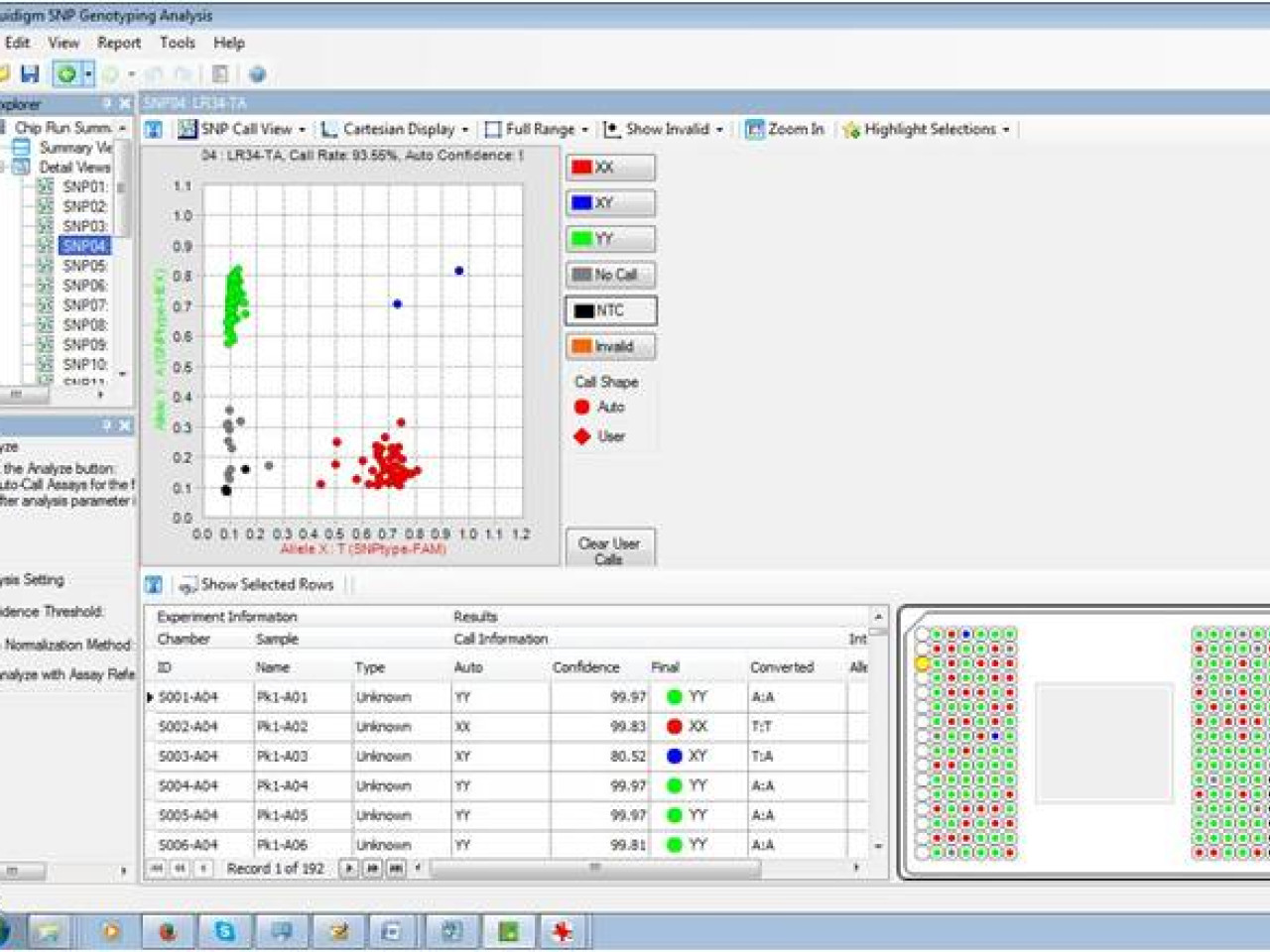 Result obtained with the developed chip
Result obtained with the developed chip

