Development of a set of 24 SNP markers (single nucleotide polymorphism) dedicated to the parallel analysis of 192 samples to test genes / markers associated with characters of interest, such as quality and low-toxicity gluten. SNP-based markers (based on differences of single nucleotides) are among the most frequently used for the molecular characterization of plant organisms. The use of EP1 platform – Fluidigm equipment, thanks to its high processivity (24 x 192 analysis in a single day) increases the efficiency of assisted selection projects that require evaluation of reeding populations made up of many individuals.
 Genotyping platform EP1
Genotyping platform EP1
SNP markers (single nucleotide polymorphism) uniformly distributed in the genomes, represent the most frequent type of genetic variation. Their advantages are given by the presence of only 2 alleles and the transferability of protocols between laboratories, experiments, etc. The EP1 analysis platform can be applied to different agri-food sectors and allows us to offer to companies a custom genotype analysis and molecular selection service quickly and at low cost.
The characteristics of the 24 x 192 chip, make the set of markers easily applicable to assited selection of cereal species (eg wheat), the varietal constitution and to the characterization of seed companies’ plant material collections. In case of specific requests of companies our laboratory is able to develop dedicated sets of 24 SNP markers for different agricultural species and to provide genotyping and assisted selection services with 1-24 (and multiples of 24) markers. The service is addressed to seed companies and to breeders.
 Biogest Siteia chip used for analysis
Biogest Siteia chip used for analysis
Characterization of a collection of wheat with the set of 24 SNP markers associated with genes coding for gluten proteins for the selection of the most suitable varieties for the production of food for individuals predisposed to celiac disease
The set of 24 markers was developed as part of the POR-FESR project at purpose of characterizing a collection of wheat useful for the study of the character "low-toxicity gluten". The study was divided into a first phase of bibliographic research for the evaluation of genes underlying the character. The next step involved a bioinformatic analysis of the sequences in order to dentifiy polymorphisms (SNP) responsible for allelic variants with improved characteristics. After the first optimization phase that led to the choice of the most suitable markers, a final set of 24 coding local-specific SNP markers was obtained.
SITEIA Parma (UNIPR) CIRI- Agroalimentare (UNIBO) Biopharmanet-Tec (UNIPR)
The data obtained with the SNP markers and more detailed phenotypic evaluations, allowed identification of the accessions with characteristics desired for the improvement of the "low-toxicity gluten" character and for the production and the wheat quality.
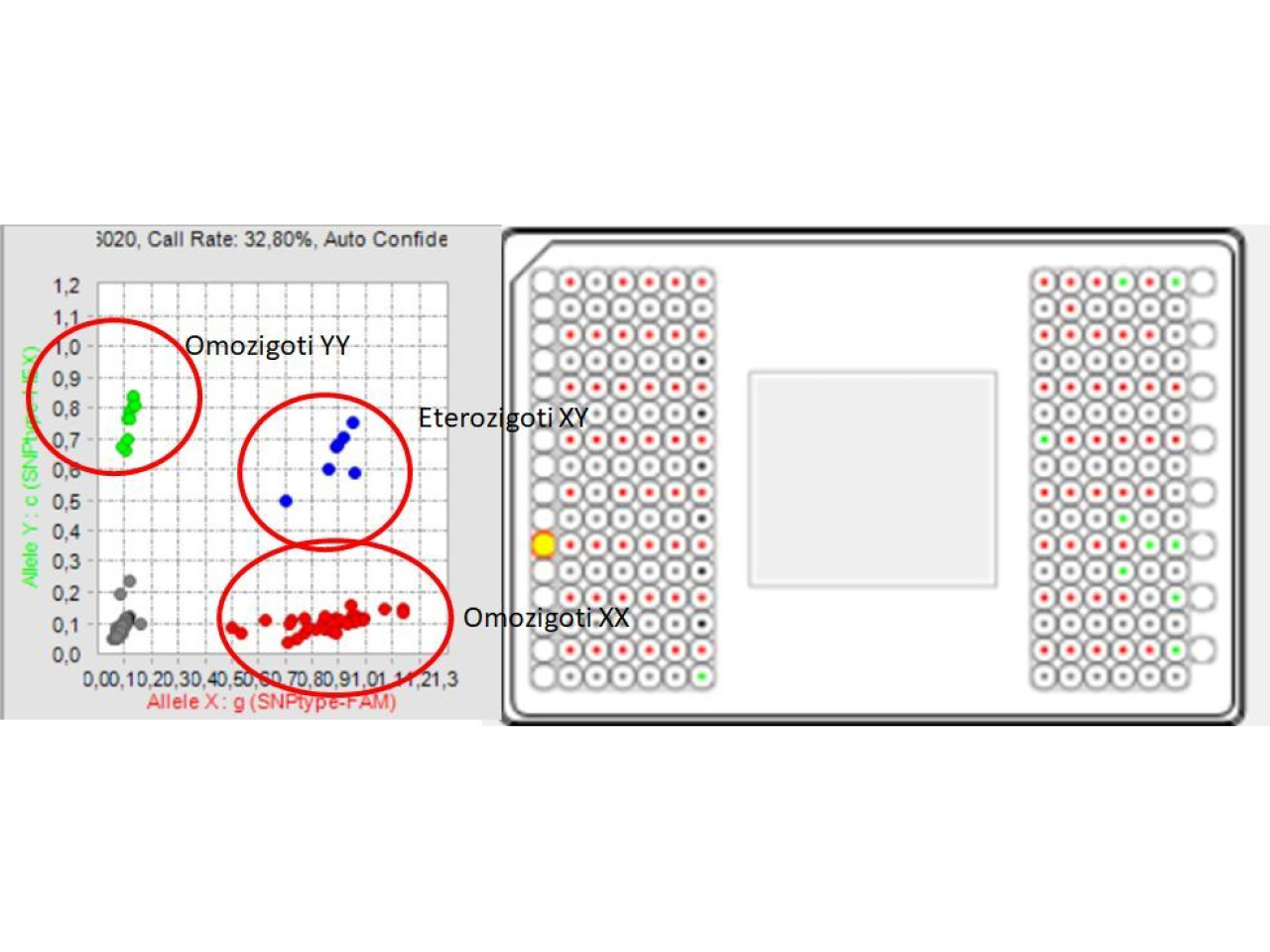 Results obtaines with the developed set of markers
Results obtaines with the developed set of markers

